Fluorosequencing is a proteomics game changer
Key Concept
With help from a reference database, only a few amino acids are needed to uniquely identify a protein in the proteome

Example: Cataloging all the proteins of a single cell
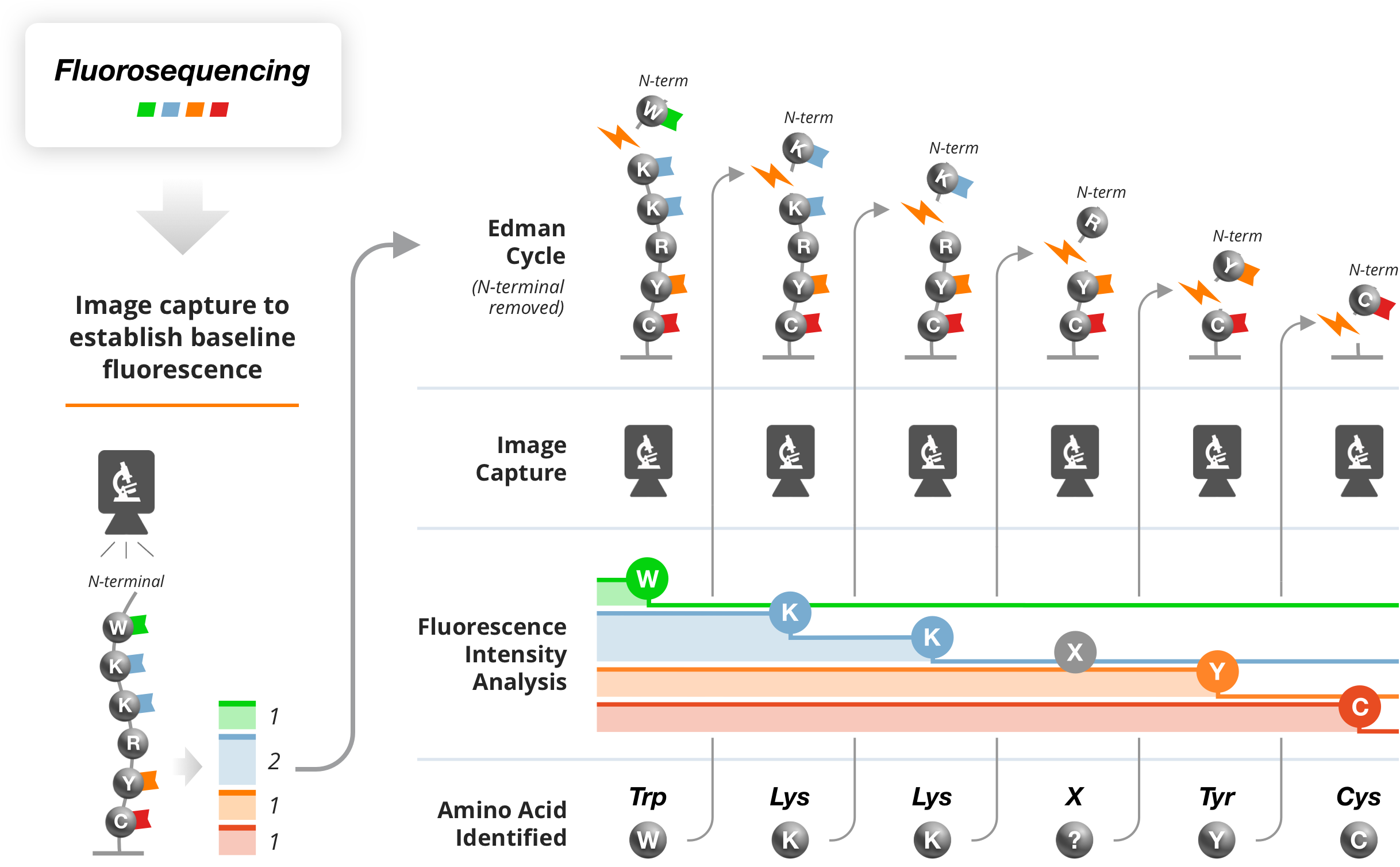
The key concept of Fluorosequencing as described in Nature Biotechnology article is as follows: First, one or more amino acid types on the peptides are selectively chemically labeled with specific identifier fluorophores. Millions of individual peptides are then immobilized on a glass cover slip. Next, each molecule’s fluorescence is monitored using total internal reflection fluorescence (TIRF) microscopy following consecutive rounds of amino-terminal (N-terminal) amino acid removal by Edman chemistry. The sequence positions of the labeled amino acids are thus identified for each peptide molecule, providing a partial sequence for each molecule. These fluorescence signatures (“fluorosequences”) can then be compared to a reference proteome for assignment to their proteins of origin.
Features and benefits
The ability to image and sequence individual peptide molecules makes the technology immediately 1 million times more sensitive than the current peptide mass- spectrometers. This offers the benefit of using extremely low sample amounts, such as clinical biopsies. This smaller sample requirements, assuming a factor of at least 1000, roughly translates to a difference between needing 1/4th of a pancreas or sampling with a needle for biopsy material.
The ability to sequence and independently measure fluorescence from individual peptide molecules tethered to an imaging surface, provides a scalable architecture to achieve a throughput to identify billions of peptide molecules on a single glass slide.
Since every single molecule measurement is independent of another, fluorosequencing platform can discriminate peptides and proteins across a large range of heterogeneity and abundances (up to 10^6 range). This means that if there is a single peptide, mixed with 1 million copies of a different peptide, the two peptides can both be identified. This helps identifying peptides existing in an extremely diverse backgrounds, such as antigens on tumor surfaces or discriminating low abundant phosphorylation event on a protein.
Unlike other proteomic technologies, where peptides and proteins are quantified relative to a standard (often using known concentrations of itself), quantification in fluorosequencing is done by counting molecular observations. This type of counting statistics makes it possible to compare abundances between proteins in the same and across experiments without need for external calibrants.
Fluorosequencing vs the competition
- Indirect measurement
- Diverse set of peptides
- Poorly correlated quantification
- Ultimate sensitivity
- Ultra-low sample requirement
- Massively parallel throughput
- Absolute quantification
- De novo PTM discovery
- High sample requirement
- Sequential processing
- PTM low resolution

